retistruct
Retistruct
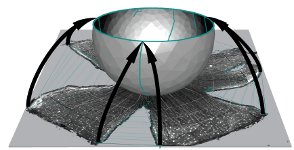 Computational reconstruction and transformation of flattened retinae
Computational reconstruction and transformation of flattened retinae
Retistruct is an R package to morph a flat surface with cuts (a dissected flat-mount retina) onto a curvilinear surface (a standard retinal shape). It can estimate the position of a point on the intact adult retina to within 8° of arc (3.6% of nasotemporal axis). The coordinates in reconstructed retinae can be transformed to visuotopic coordinates.
How Retistruct works
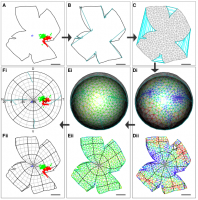
Reconstruction is achieved by: stitching the marked-up cuts of the flat-mount outline; dividing the stitched outline into a mesh whose vertices then are mapped onto a curtailed sphere; and finally moving the vertices so as to minimise a physically-inspired deformation energy function.
- Full details are in the paper: Sterratt, D. C., Lyngholm, D., Willshaw, D. J. and Thompson, I. D. (2013). Standard Anatomical and Visual Space for the Mouse Retina: Computational Reconstruction and Transformation of Flattened Retinae with the Retistruct Package. PLoS Computational Biology 9:e1002921.
- This poster and this YouTube video, presented at the 2012 Neuroinformatics Meeting in Munich, has more details and examples of reconstructions and projections into visual space.
Installation and documentation
Retistruct has been tested on GNU/Linux, Mac OS X and Microsoft Windows.
Stable version
To install the stable version of Retistruct hosted on CRAN, follow the instructions in the User Guide. The installation contains a number of demonstration retinae, and instructions for how to handle retinal flat-mount images in Retistruct.
Development version and source code
The development version of Retistruct contains the most recent bug fixes and improvements, but may not be stable. To install it follow the instructions on the Retistruct Github repository, where the source code can also be checked out.
Reference publication
For reference purposes, this zip file contains the version of Retistruct that generated the reconstructions in Sterratt & al. (2013; PLoS Computational Biology 9). The file also contains some Matlab code to read data directories contained by Retistruct.
Sample data
As well as the built-in demo data, there are some sample images to practise on:
- Beginner: SMI-32 stained retina. As described in the User Guide, the outline can be marked up in ImageJ and imported into Retistruct.
- More advanced: TIFF files (left and right), each containing a stack of three images corresponding to Figure 6 of the manuscript: retinae labelled with Fluoro-Emerald, Fluoro-Ruby and a brightfield image. As described in the User Guide, use ImageJ to mark up the outline on the brightfield image, and then use ImageJ’s particle analyser to find the locations of the stained cells.
Problems?
If you encounter issues using Retistruct please either:
Work using Retistruct
- Gupta D., Młynarski W., Sumser A., Symonova O., Svatoň J., and Joesch M. (2023). “Panoramic visual statistics shape retina-wide organization of receptive fields”. Nat. Neurosci. 26: 606–614 DOI:10.1038/s41593-023-01280-0
- D’Souza, S. P., Swygart, D. I., Wienbar, S. R., Upton, B. A., Zhang, K. X., Mackin, R. D., Casasent, A. K., Samuel, M. A., Schwartz, G. W. and Lang, R. A. (2022). “Retinal patterns and the cellular repertoire of neuropsin (Opn5) retinal ganglion cells”. J. Comp. Neurol. 530: 1247-1262. DOI: 10.1002/cne.25272
- Tosetto, L., Williamson, J. E., White, T. E. and Hart, N. S. (2021) “Can the Dynamic Colouration and Patterning of Bluelined Goatfish (Mullidae; Upeneichthyslineatus) Be Perceived by Conspecifics?”. Brain Behav. Evol. DOI: 10.1159/000519894
- Adelman, S. A., Oikawa, K., Senthilkumar, G., Trane, R. M., Teixeira, L., and McLellan, G. J. (2021). “Mapping retinal ganglion cell somas in a large-eyed glaucoma model”. Mol. Vis. 27: 608–621. PMC
- Nadal-Nicolás, F. M., Kunze, V. P., Ball, J. M, Peng, B. T., Krishnan, A., Zhou, G., Dong, L. and Li, W. (2020). “True S-cones are concentrated in the ventral mouse retina and wired for color detection in the upper visual field”. eLife 9: e56840 DOI: 10.7554/eLife.56840
- Deichler, A., Carrasco, D., Lopez-Jury, L. et al. (2020). “A specialized reciprocal connectivity suggests a link between the mechanisms by which the superior colliculus and parabigeminal nucleus produce defensive behaviors in rodents”. Sci. Rep. 10: 16220 DOI: 10.1038/s41598-020-72848-0
- Lyngholm, D., Sterratt, D. C., Hjorth, J. J., Willshaw, D. J., Eglen, S. J., and Thompson, I. D. (2019). “Measuring and Modelling the Emergence of Order in the Mouse Retinocollicular Projection”. bioRxiv: 713628. DOI: 10.1101/713628
- Sondereker, K. B., Stabio, M. E., Jamil, J. R., Tarchick, M. J. and Renna, J. M. (2018) “Where You Cut Matters: A Dissection and Analysis Guide for the Spatial Orientation of the Mouse Retina from Ocular Landmarks”. J Vis. Exp. 138 DOI: 10.3791/57861
- Stabio, M. E., Sondereker K. B., Haghgou S. D., Day, B. L., Chidsey, B., Sabbah, S. and Renna, J. M. (2018) “A novel map of the mouse eye for orienting retinal topography in anatomical space”. J. Comp. Neurol. 526:1749-1759. DOI: 10.1002/cne.24446
- Smodlaka, H., Khamas, W. A., Palmer, L., Lui, B., Borovac, J. A., Cohn, B. A. and Schmitz, L. (2016) “Eye Histology and Ganglion Cell Topography of Northern Elephant Seals (Mirounga angustirostris)” Anat. Rec. (Hoboken) 6: 798-805. DOI: 10.1002/ar.23342
- Cohn, B. A., Collin, S. P., Wainwright, P. C. and Schmitz, L. (2015) “Retinal topography maps in R: new tools for the analysis and visualization of spatial retinal data”. J. Vis. 15: 19. DOI: 10.1167/15.9.19
- Flinn, J. M., Kakalec P., Tappero, R., Jones, B. and Lengyel, I. (2014) “Correlations in distribution and concentration of calcium, copper and iron with zinc in isolated extracellular deposits associated with age-related macular degeneration”. Metallomics 6: 1223-1228. DOI: 10.1039/c4mt00058g
- Bleckert, A., Schwartz, G. W., Turner, M.H., Rieke, F. and Wong, R. O. L. (2014) “Visual space is represented by nonmatching topographies of distinct mouse retinal ganglion cell types”. Current Biology 24: 310-315. DOI: 10.1016/j.cub.2013.12.020
Updates
- August 2020
- The development branch, 0.7.x, is released. This adds the ability to stitch together separate fragments and reconstruct 3D data comprising an overhead image and depth map. There are also user interface improvements, most notably a magnifier function. The improvements were supported by an NIH R21 grant (EY027894) from 2018-2020 to Dr. Mark P. Krebs, The Jackson Laboratory.
- December 2019
- Retistruct 0.6.1 and 0.6.2 is out. This reinstates sphericalplots and the saving of reconstructed data. New features include allowing resizing of windows so plots can be larger, allowing data counts to be read in using ijroi and csv formats and exposing some features in the API. Version 0.6.2 fixes a small issue and adds much better documentation for developers. See the NEWS file for more information.
- July 2019
- Retistruct 0.6.0 is out. The big change in this version is a complete refactor of the code, which should enable future enhancements to happen more quickly. The stitching algorithm is improved. There are major improvements in image rendering, which should be a lot faster thanks to the tsearch algorithm in 0.4.x of the geometry package, and able to deal with high-resolution images.
- July 2017
- Retistruct 0.5.11 is out. The CSV input format can now take a file of counts (or “grouped”) data and there are a number of bug fixes; see the NEWS file for more information.
- March 2015
- Brian Cohn has pointed out a precursor paper to Retistruct, which we
weren’t aware of when developing Retistruct, or writing the paper:
- Curcio, C.A., Sloan, K. R. and Meyers, D. (1989) “Computer methods for sampling, reconstruction, display and analysis of retinal whole mounts”. Vision Res. 29(5):529-40. Pubmed
Authors and funding
Retistruct was written by David Sterratt at the University of Edinburgh, and tested by Daniel Lyngholm and Ian Thompson at the MRC Centre for Developmental Neurobiology, KCL.
The development of the initial version of Retistruct was supported by a Programme Grant from the UK Wellcome Trust (G083305) from 2008-2013.
Improvements to image handing and refactoring the code (released in v0.6.0) were supported by The Jackson Laboratory (Bar Harbor, ME, USA) Scientific Services Innovation Fund from 2016-2017 and an NIH R21 grant (EY027894) from 2018-2020 to Dr. Mark P. Krebs, The Jackson Laboratory.
The capabilities to reconstruct tissue comprised of separate fragments (released in v0.7.0) and to reconstruct 3D data comprising an overhead image and depth map (released in v0.7.2), and user interface improvements (released in v0.7.0) were supported by an NIH R21 grant (EY027894) from 2018-2020 to Dr. Mark P. Krebs, The Jackson Laboratory.
–>